Let's perform a viral phylogenetic analysis on HIV-1 genomes
Now you have the tools you need to perform a full phylogenetic analysis! Let’s take a look at a slightly more challenging dataset: HIV-1 whole genome sequences collected from real people. Based on our dataset of 10 HIV-1 sequences, can you accurately date the MRCA?
Take the time to go through the same set of steps as you did with SARS-CoV-2:
Run multiple sequence alignment with ViralMSA using
hiv1_sequences.fasandhiv_reference.fas.Generate an unrooted tree with FastTree
Generate a rooted tree with LSD2 using
hiv1_dates.textandhiv1_outgroups.txt
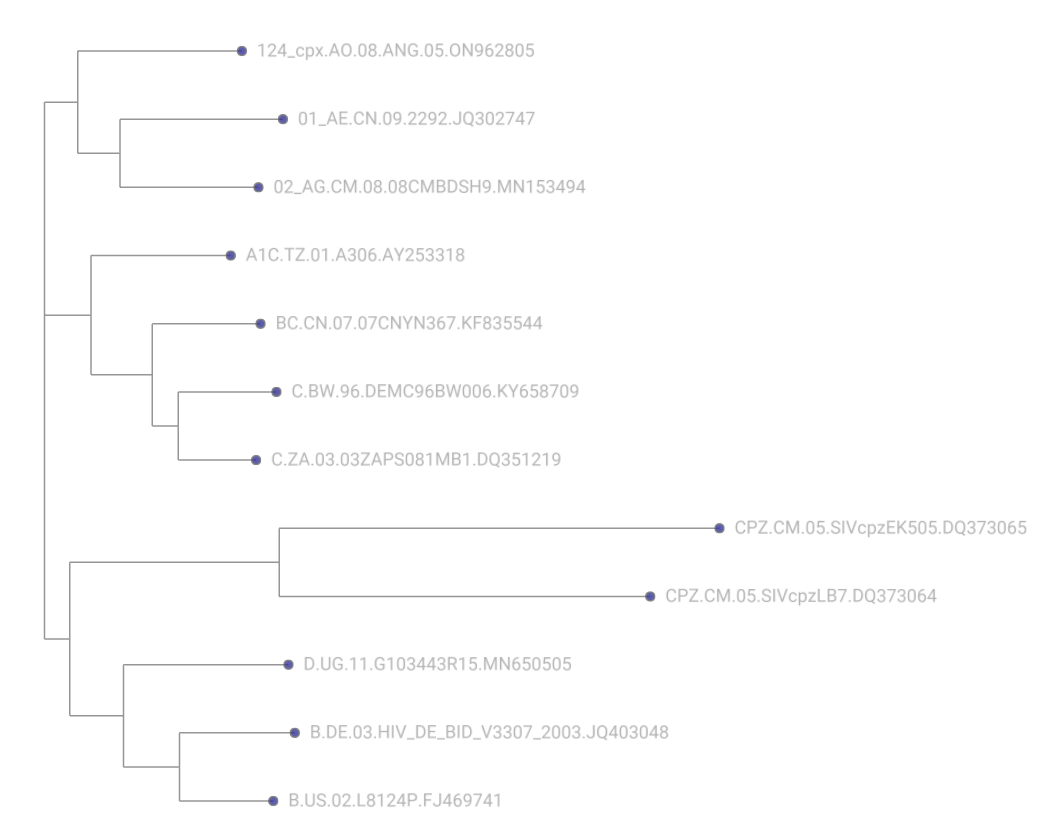
Now, let’s take a look at the trees to ensure that they reflect what we would expect from the data. We have provided both the unrooted and rooted trees below so that you can compare your results:
Unrooted
Rooted
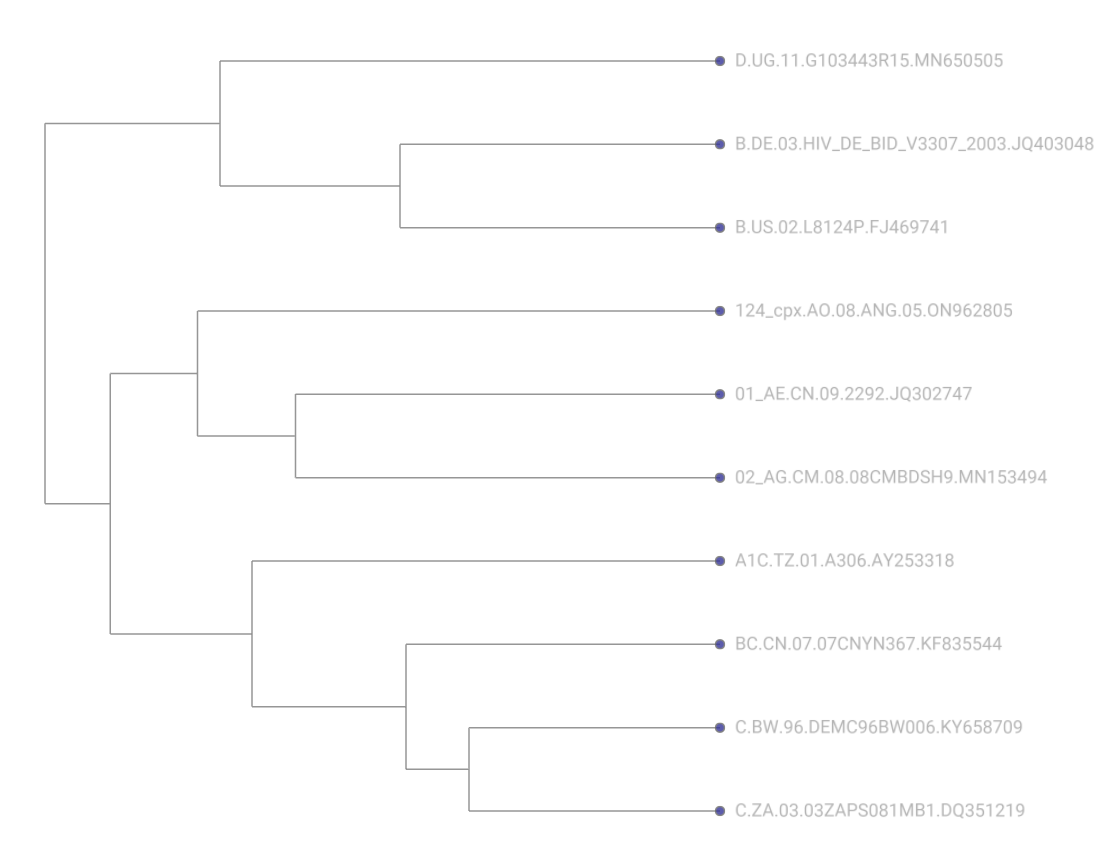
Take a look at the log file from LSD2. Does the date of the MRCA seem reasonable?
You may find that the date of the MRCA doesn’t make the most sense! This highlights the difficulty of phylogenetic rooting and dating and the need to have not only a high-quality initial sequence dataset, but also to understand the computational tools you use to perform the analysis as modification of parameters (or a different tool altogether!) may be more appropriate.