No doubt readers familiar with BLAST have been curious: aren’t there databases of some kind involved in BLAST searches? Not necessarily. Simple FASTA files will suffice for both the query and subject set. It turns out, however, that from a computational perspective, simple FASTA files are not easily searched.
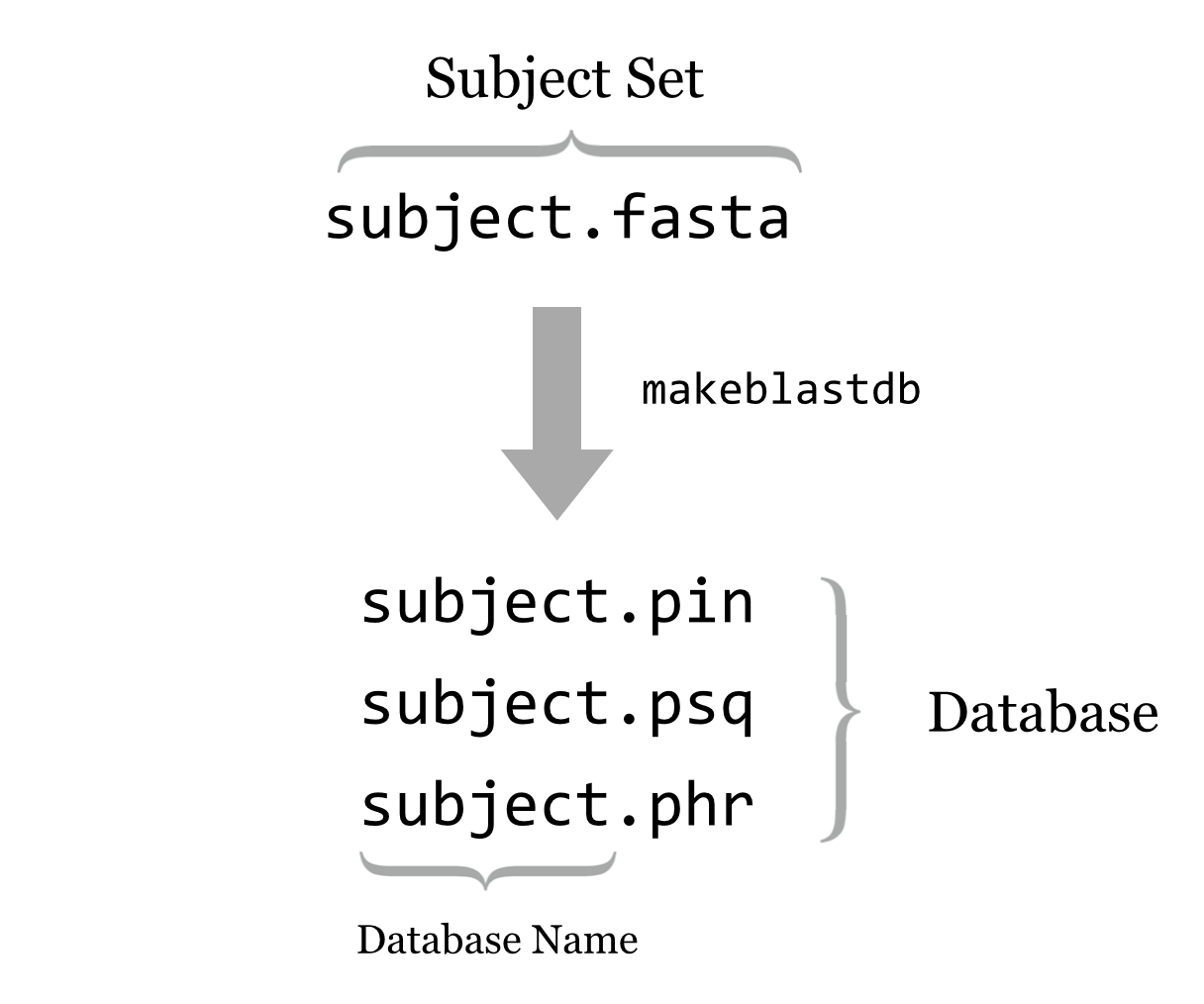
Thus BLAST+ provides a tool called makeblastdb that converts a subject FASTA file into an indexed and quickly searchable (but not human-readable) version of the same information, stored in a set of similarly named files (often at least three ending in .pin, .psq, and .phr for protein sequences, and .nin, .nsq, and .nhr for nucleotide sequences).
This set of files represents the “database,” and the database name is the shared file name prefix of these files: