BLAST is not a single tool, but rather a suite of tools (and the suite grows over the years as more features and related tools are added). The most modern version of the software, called BLAST+, is maintained by the NCBI.
The programs in the BLAST+ suite can search for and against sequences in protein format and in nucleotide format (A’s, C’s, T’s, and G’s). Depending on what type the query and subject sets are, different BLAST programs are used:
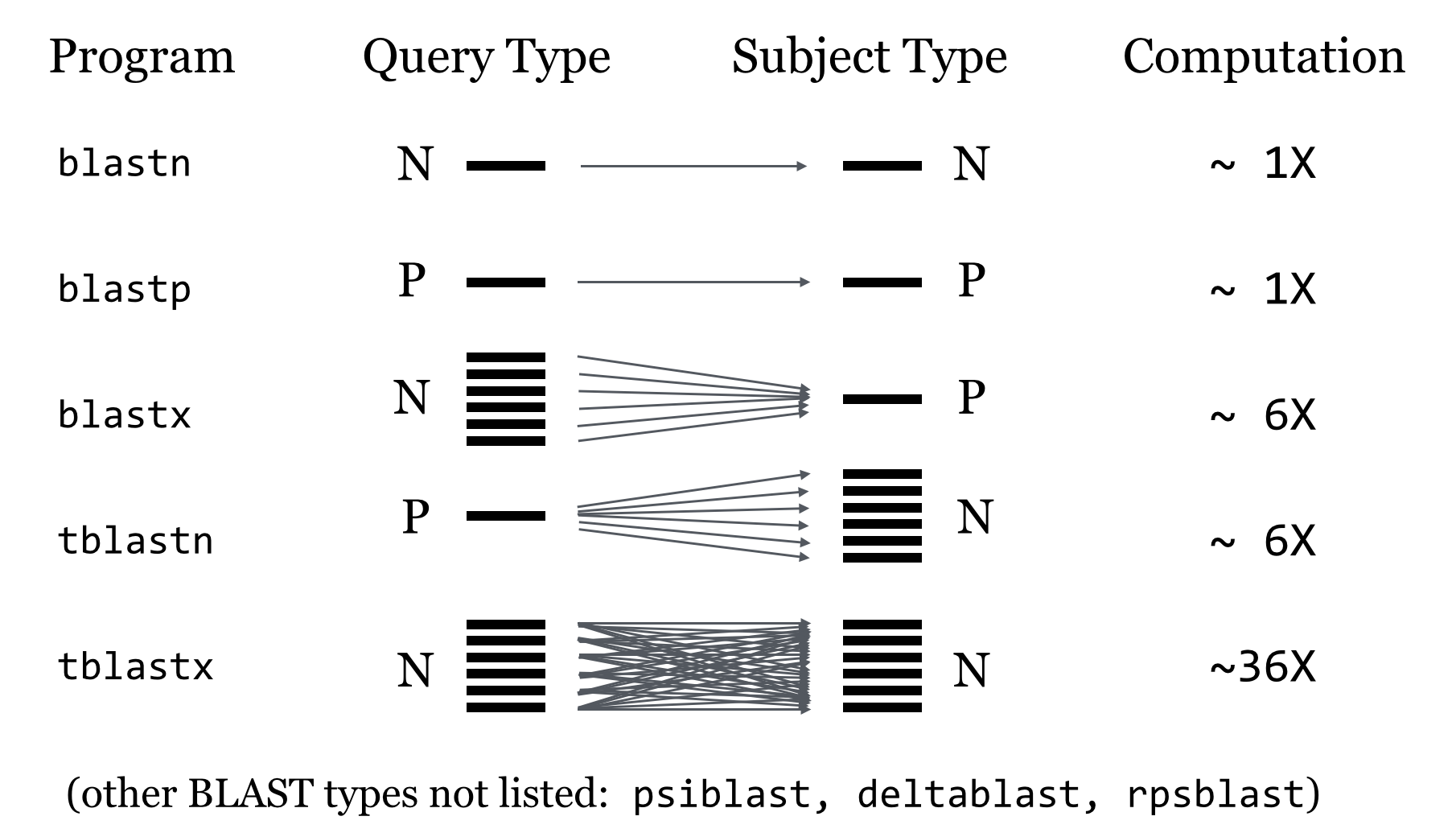
While two nucleotide sequences (N comparisons in the figure above) may be compared directly (as may two protein sequences, represented by P), when we wish to compare a nucleotide sequence to a protein sequence, we need to consider which reading frame of the nucleotide sequence corresponds to a protein.
The blastx and tblastn programs do this by converting nucleotide sequences into protein sequences in all six reading frames (three on the forward DNA strand and three on the reverse) and comparing against all of them. Generally such programs result in six times as much work to be done.
The tblastx program compares nucleotide queries against nucleotide subjects, but it does so in protein space with all six conversions compared to all six on both sides.