Bedtools tutorial
Help 7 / 20
bedtools intersect
Faster with sorted data
So far the examples presented have used the traditional algorithm in bedtools for finding intersections. It turns out, however, that bedtools is much faster when using presorted data.
For example, compare the difference in speed between the two approaches when finding intersections between exons.bed and hesc.chromHmm.bed:
time bedtools intersect -a gwas.bed -b hesc.chromHmm.bed > /dev/null
time bedtools intersect -a gwas.bed -b hesc.chromHmm.bed -sorted > /dev/null
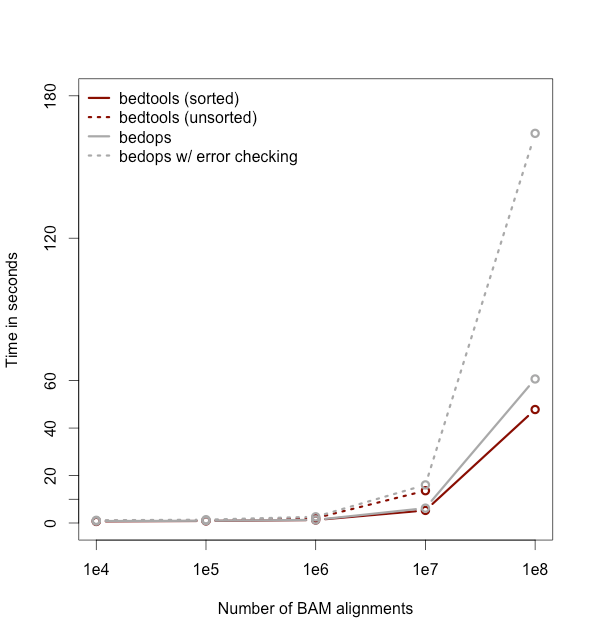
Note: While the run times in this example are quite small, the performance gains from using the -sorted option grow as datasets grow larger. For example, compare the runtimes of the sorted and unsorted approaches as a function of dataset size in the figure below.
The important thing to remember is that each dataset must be sorted by chromosome and then by start position: sort -k1,1 -k2,2n.
Loading...